The Data Science Checklist: Best Practices for Maintainable Data Science Projects
Over the years, I’ve developed dozens of Data Science and Machine Learning projects, and I’ve learned a few lessons the hard way. Without structure and standards, chaos takes over, making it challenging to make consistent progress. I’ve compiled a list of best practices to ensure projects and maintainable, and I’m sharing this with you so you can use it for your next project. Let’s get started!
Summary
Before we dive into the details on each item, here’s the summary:
- Store data, artifacts, and code in different locations
- Declare paths and configuration parameters in a single file
- Store credentials securely
- Keep your repository clean by using a
.gitignorefile - Automate the workflow end-to-end
- Declare all software dependencies
- Split production and development dependencies
- Have an informative and concise README
- Document code
- Organize your repository in a hierarchical structure
- Package your code
- Keep your Jupyter notebooks simple
- Use the logging module (do not use print)
- Test your code
- Take care of code quality
- Delete dead and unreachable code
1. Store data, artifacts, and code in different locations
When working on a project, we’ll encounter three types of files:
- Data. All the raw data and any intermediate data results (e.g., CSV, txt, SQL dumps, etc.)
- Artifacts. Any other files generated during a pipeline execution (e.g., a serialized model, an HTML report with model metrics)
- Source code. All the files to support the pipeline execution (e.g.,
.py,.Rsource code, Jupyter notebooks, tests)
To keep things organized, separate each group into different folders; doing so will make it easier to understand the project structure:
# YES: separate data and artifacts from code
data/
raw.csv
...
artifacts/
model.pkl
report.html
...
src/
download_raw.py
clean.py
featurize.py
fit.py
...
# NO: mix data, artifacts and code
myproject/
raw.csv
model.pkl
report.html
download_raw.py
clean.py
featurize.py
fit.py
...
2. Declare paths and configuration parameters in a single file
Your pipeline needs to load its raw data from a particular source; furthermore, you may need to interact with other systems (such as a data warehouse). Finally, you need to store results (either the local filesystem or remote storage). A common anti-pattern is embedding these configuration parameters in the source code, leading to the following issues:
- Ambiguity. Not disclosing external systems hides essential implementation details, which can confuse collaborators: where is the data located? Does this project need access to the Kubernetes cluster?
- Opacity. If there is no single place to store temporary files, each collaborator may start keeping outputs in arbitrary locations, making it difficult to organize the project.
- Redundancy. Infrastructure resources (i.e. data warehouse, Kubernetes cluster) are likely to be used multiple times, leading to duplicated configuration if the configuration parameters are not centralized.
YES: centralize paths and configuration parameters
# config.yaml - located in the root folder
path:
raw: /path/to/raw/
processed: /path/to/processed/
artifacts: /path/to/artifacts/
infrastructure:
# important: do not include any
# credentials (such as db passwords) here!
cluster: cluster.my_company.com
warehouse: warehouse.my_company.com
…then read from there:
# some_task.py
from pathlib import Path
# assume you created a load_config data that reads your config.yaml
from my_project import load_config
import pandas as pd
cfg = load_config()
raw_data = pd.read_parquet(Path(cfg['raw'], 'data.parquet'))
# do things...
path_to_report = Path(cfg['artifacts'], 'report', 'lastest.html')
path_to_report.write_text(report)
NO: hardcoded values will make your life harder
import pandas as pd
# hardcoded path/to/raw!
raw_data = pd.read_parquet('pat/to/raw/data.parquet')
3. Store credentials securely
You should never store database credentials (or any other type of credentials) in plain text files (that includes .py source code or any configuration files).
YES: Securely store your credentials (this example uses the keyring library)
from my_project import load_locations
import psycopg2
import keyring
def open_db_conn():
"""Open a db connection
Notes
-----
You have to store the credentials before running this function:
>>> keyring.set_password("mydb", "username", "mypassword")
"""
loc = load_locations()
db = loc["servers"]["database"]
dbname = db["name"]
user = db["username"]
host = db["host"]
# securely retrieve db password
password = keyring.get_password(dbname, user)
conn = psycopg2.connect(dbname=dbname,
host=host,
user=user,
password=password)
return conn
Then in your scripts:
from my_project import open_db_conn
# securely load credentials, then open db connection
dbname, host, user, password = open_db_conn()
# do some stuff with the data
The keyring uses your OS credential store, safer than a plain text file. The previous example only stores the password, but you can store any of the other fields.
NO: credentials stored in a source file
import psycopg2
conn = psycopg2.connect(dbname='dbname',
host='data.company.com',
user='username',
# do not store passwords in plain text
password='mypassword')
4. Keep your repository clean by using a .gitignore file
Most likely, you’re storing your code in a git repository. Therefore, it is essential to specify a .gitignore file to keep it clean. Missing this file can make your repository unnecessarily messy and might break the project for your colleagues (i.e. if you commit some user-dependent configuration file).
What to include in the .gitignore file? First, grab the Python template from Github, which contains pretty much what you need. Then make sure you also include common extensions to avoid accidentally committing data or configuration files. For example, if your raw data comes in CSV/JSON formats and you have some user-dependent YAML configuration files, add the following lines:
# data files
*.csv
*.json
# configuration files
*.yaml
If you have already committed some of those files, you can remove them using the following commands:
git rm -r --cached .
git add .
git commit -m 'Applies new .gitignore'
Note that this will not remove files from previous commits. Doing so from the git is CLI, but the BFG tool simplifies the process.
5. Automate the data workflow end to end
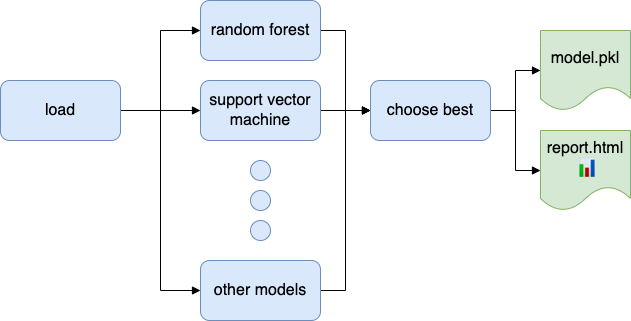
A data project usually requires multiple data transformations. Therefore, automating the entire process is essential to ensure reproducible code. Not having a fully automated analysis will cause trouble down the road. For example, say you have to re-train a model in a project written by someone else, so you see the following line in the README.md:
# train a model and export a report with results
python -m otherproject.model --type=random_forest --report=my_model.html
But then get an error like this:
SQL Error [42P01]: ERROR: relation "training_set" does not exist
Well, it makes sense that you should build features before attempting to train a model, but at this moment, you will be wondering how to do so.
Ploomber makes it simple to automate data workflows, you can add as many scripts or notebooks as you want, then orchestrate execution with:
ploomber build
6. Declare software dependencies
Data Science projects usually depend on third-party packages (e.g. pandas, scikit-learn); failing to specify them will cause trouble to any new collaborator. How many times have you seen this error?
ModuleNotFoundError: No module named 'X'
Although the error message is very descriptive, it is a great practice to document all the dependencies so anyone can quickly get started. In Python, you can achieve this via a requirements.txt file, which contains the name of a package in each line. For example:
pandas
scikit-learn
Then, you can install all dependencies with:
pip install -r requirements.txt
If you are using Ploomber, you can use the following command that will take care of setting up a virtual environment and installing all the declared dependencies:
ploomber install
7. Split production and development dependencies
It’s good to keep the number of dependencies at a minimum. The more dependencies your project needs, the higher the chance of running into installation issues. A way to simplify installation is to split our dependencies into two groups:
- Packages required to run the pipeline - These are the packages needed to run the pipeline in production.
- Optional packages required to develop the pipeline - These are optional packages you need to work on the project (e.g., building documentation, running exploratory Jupyter notebooks, running tests, etc.) but aren’t needed in production.
For our case, you might want two keep two dependency files (say requirements.prod.txt and requirements.dev.txt). Then, during development, you can run:
pip install -r requirements.dev.txt
And in production:
pip install -r requirements.prod.txt
Important: When running in production, it’s essential to declare specific versions of each package; you can use the pip freeze command to export a file with particular versions of each package installed. If you want to save the trouble, you may use Ploomber and execute ploomber install, which will store a separate requirements.txt (dev and prod) with specific versions of each package.
8. Have an informative and concise README
The README file is the entry point for anyone using the pipeline; it is essential to have an informative one to help collaborators understand how the source files fit together.
What to include in the README file? Basic information about the project, at the very least, include a brief description of the project (what does it do?), list data sources (where does the data come from?), and instructions for running the pipeline end-to-end.
Example:
### Boston house value estimation project
This project creates a model to predict the value of a house using the Boston housing dataset: https://www.kaggle.com/c/boston-housing
#### Running the pipeline
```sh
# clone the repo
git clone http://github.com/edublancas/boston-housing-project
# install dependencies
pip install -r requirements.txt
# run the pipeline
python run.py
```
9. Code documentation
As new contributors join the project, documentation becomes more important. Good documentation assists in understanding how the different pieces fit together. Furthermore, documentation will be helpful for you to quickly get the context of the code you wrote a while ago.
But documentation is a double-edged sword; it will cause trouble if outdated. So, as a general recommendation, keep your code simple (good variable names, small functions), so others can understand it quickly and have some minimal documentation to support it.
How to document code? Add a string right next to the function definition (aka a docstring) to provide a high-level description and convey as much information as possible using the code itself.
def remove_outliers(feature):
"""Remove outliers from pandas.Series by using the interquartile range
Parameters
----------
feature : pandas.Series
Series to remove outliers from
Returns
-------
pandas.Series
Series without outliers
"""
# actual code for computing features
pass
There are many docstring formats you can choose from, but the standard format in the Python scientific community (used in the example above) is numpydoc.
Once you added a docstring, you can quickly access it from Jupyter/IPython like this:
# running this on Jupyter/IPython
# (does not work with the standard python interpreter)
?remove_outliers
# will print this...
# Remove outliers from pandas.Series by using the IQR rule
# Parameters
# ----------
# feature : pandas.Series
# Feature to clean
# Returns
# -------
# pandas.Series
# Clean version of the feature
# Examples
# --------
# >>> clean = remove_outliers(feature)
# File: ~/path//<ipython-input-1-d0cbf0151fd4>
# Type: function
10. Organize your repository in a hierarchical structure
As your project progresses, your codebase will grow. You will have code for exploring data, loading, generating features, training models, etc. It is a good practice to spread out the code in multiple files, and organization of those files is essential; take a look at the following example:
README.md
.gitignore
exploration-dataset-a.ipynb
exploration-dataset-b.ipynb
db.py
load.py
load_util.py
clean.py
clean_util.py
features.py
features_util.py
model.py
model_util.py
The flat structure makes it hard to understand what is going on and how files relate to each other. Therefore, it is better to have a hierarchical structure like this:
README.md
.gitignore
# exploratory notebooks
exploration/
dataset-a.ipynb
dataset-b.ipynb
# utility functions
src/
db.py
load.py
clean.py
features.py
model.py
# steps in your analysis
steps/
load.py
clean.py
features.py
model.py
11. Package your code
Whenever you run pip install, Python makes the necessary configuration adjustments to ensure the package is available to the interpreter so you can use it in any Python session using import statements. You can achieve the same functionality for your utility functions by creating a Python package; this gives you great flexibility to organize your code and then import it anywhere.
Creating a package is just adding a setup.py file. For example, if your setup.py exists under my-project/ you will be able to install it like this:
pip install --editable my-project/
Note: We pass the --editable flag for Python to reload the package contents, so you’ll use the latest code whenever you update your code and start a session.
Once the package is installed, you can import it like any other package:
from example_pkg import some_function
some_function()
If you modify some_function, you will have to restart your session to see your changes (assuming you install the package in editable mode), if using IPython or Jupyter, you can avoid having session restart by using the autoreload:
%load_ext autoreload
%autoreload 2
from example_pkg import some_function
some_function()
# edit some_function source code...
# this time it will run the updated code
some_function()
For a more detailed treatment on Python packaging, see our blog post.
12. Keep your Jupyter notebooks simple
Notebooks are a fantastic way of interacting with data, but they can get out of hand quickly. If not taken proper care of, you may copy-paste code among notebooks and create big monolithic notebooks that break. So we wrote an entire article on writing clean notebooks check it out..
13. Use the logging module (do not use print)
Logging is a fundamental practice in software engineering; it makes diagnosing and debugging much more manageable. The following example shows a typical data processing script:
import pandas as pd
df = pd.read_csv('data.csv')
# run some code to remove outliers
print('Deleted: {} outliers out of {} rows'.format(n_outliers, n_rows))
The problem with the example above is using the print statement. print sends a stream to standard output by default. So if you have a few print statements on every file, your terminal will print out dozens and dozens of messages every time you run your pipeline. The more print statements you have, the more complex it is to see through the noise, and they’ll end up being meaningless messages.
Use the logging module instead. Among logging’s most relevant features are: filtering messages by severity, adding a timestamp to each record, including the filename and the line where the logging call originated, among others. The example below shows its simplest usage:
import logging
import pandas as pd
logger = logging.getLogger(__name__)
logging.basicConfig(filename='my.log', level=logging.INFO)
df = pd.read_csv('data.csv')
# remove outliers
logger.info('Deleted: {} outliers out of {} rows'.format(n_outliers, n_rows))
Note that Using the logging requires some configuration. Check out the basic tutorial.
14. Test your code
Testing data pipelines is challenging, but it is a good time investment as it pays in the long run since it allows you to iterate faster. While it is true that a lot of the code will change during the experimental phase of your project (possibly making your tests outdated), testing will help you make more consistent progress.
We have to consider that Data Science projects are experimental, and the first goal is to see if the project is even is feasible, so you want to get an idea as soon as possible. But, on the other hand, a complete lack of testing might get you to the wrong conclusion, so you must find a balance.
At a minimum, ensure that your pipeline runs end-to-end every time you merge to the main branch. We’ve extensively written on testing for data projects. Check out our article that focuses on testing Data Science projects and our article covering specifics of testing Machine Learning projects.
15. Take care of code quality
People read code more often than they write it. To make your code more readable, use a style guide.
Style guides are about consistency; they establish a standard set of rules to improve readability. Such rules have to do with maximum line length, whitespace, or variable naming. The official style guide for Python is called PEP8; if you open the link, you will notice that it is a lengthy document. Fortunately, there are better ways to get you up to speed.
pycodestyle is a tool that automatically checks your code against PEP8’s rules. It will scan your source files and show you which lines of code do not comply with PEP8. While pycodestyle limits finding PEP8 non-compliant lines to improve readability, there are more general tools.
pyflakes is a tool that also checks for possible runtime errors; it can find syntax errors or undefined variables.
If you want to use both pycodestyle and pyflakes, I suggest you use flake8, which combines both plus another tool to check for code complexity (a metric related to the number of linearly independent paths).
You can use all the previous tools via their command-line interface. For example, once flake8 is installed, you can check a file via flake8 myfile.py. To automate this process, text editors and IDEs usually provide plugins that automatically run these tools when you edit or save a file and highlight non-compliant lines. Here are instructions for VSCode.
Automatic formatting
As a complement to style linters, auto-formatters allow you to fix non-compliant lines. Consider using black. Auto-formatters can fix most of the issues with your code to make it compliant; however, there still might be things that you need to fix manually, but they’re huge time savers.
Other linters
There are other linters available; one of the most popular ones is pylint, it is similar to flake8, but it provides a more sophisticated analysis. In addition, there are some other specific tools, such as bandit, which focus on finding security issues (e.g., hardcoded passwords). Finally, if you are interested in learning more, check out the Python Code Quality Authority website on Github.
16. Delete dead and unreachable code
Make a habit of removing dead (code whose outputs are never used) and unreachable code (code which is never executed).
An example of dead code happens when we call a function but we never use their output:
def process_data(data):
stats = compute_statistics(data)
pivoted = pivot(data)
# we are not using the stats variable!
return transform(pivoted)
In our previous example, compute_statistics is dead code since we’re never using the output (stats). So let’s remove it:
def process_data(data):
pivoted = pivot(data)
return transform(pivoted)
We got rid of the stats = compute_statistics(data), but we should check if we’re calling compute_statistics anywhere else. If not, we should remove its definition.
It is essential to delete the code on the spot because the more time it passe, the more complex it is to find these kinds of issues. Some libraries help detect dead code, but it’s better to do it right when we notice it.
Final comments
I hope these suggestions help you improve your workflow. Once you get used to them, you will see that keeping your codebase maintainable increases your productivity in the long run. Ping us on Slack if you have any comments or questions.
Deploy a free JupyterLab instance with Ploomber
Try Ploomber Cloud Now
Get Started


