The did you upload the latest data version yet? nightmare
Data pipelines are multi-stage processes. Whether you are doing data visualization or training a Machine Learning model, there is an inherent workflow structure. The following diagram shows a typical Machine Learning pipeline:
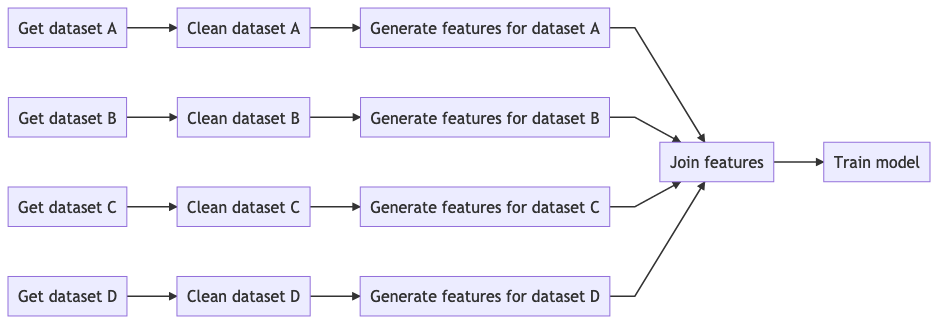
Imagine you are working with three colleagues, taking one feature branch each (A to D). To execute your pipeline, you could write a master script that runs all tasks from left to right.
During development, end-to-end runs are rare because they take too long. If you want to skip redundant computations (tasks whose source code has not changed), you could open the master script and manually run outdated tasks, but this will soon turn into a mess. Not only you have to keep track of task’s status along your feature branch, but also in every other task that merges with any of your tasks. For example, if you need to use the output from “Join features”, you have to ensure the output generated by all four branches is updated.
Since full runs take too long and keeping track of outdated tasks manually is a laborious process, you might resort to the evil trick of sharing intermediate results. Everyone uploads the latest version of all or some selected tasks (most likely the ones with the computed features) to a shared location that you can then copy to your local workspace.
But to generate the latest version of each task, you have to ensure it was generated from the latest version of all its upstream dependencies, which take us back to original problem; there are simply no guarantees about data lineage. Using a data file whose origin is unknown as input severely compromises reproducibility.
Introducing the Ploomber Spec API
The new API offers a simple way to sync Data Science teamwork. All you have to do is list source code location and products (files or database tables/views) for each task in a pipeline.yaml file.
# pipeline.yaml
# clean data from the raw table
- source: clean.sql
product: clean_data
# function that returns a db client
client: db.get_client
# aggregate clean data
- source: aggregate.sql
product: agg_data
client: db.get_client
# dump data to a csv file
- class: SQLDump
source: dump_agg_data.sql
product: output/data.csv
client: db.get_client
# visualize data from csv file
- source: plot.py
product:
# where to save the executed notebook
nb: output/executed-notebook-plot.ipynb
# tasks can generate other outputs
data: output/some_data.csv
Ploomber will analyze your source code to determine dependencies and skip a task if its source code (and the source code of all its upstream dependencies) has not changed since the last run.
Say your colleagues updated a few tasks. To bring the pipeline up-to-date:
git pull
ploomber entry pipeline.yaml
If you run the command again, nothing will be executed.
Apart from helping you sync with your team. Ploomber is great for developing pipelines iteratively: modify any part and call build again, only modified tasks will be executed. Since the tool is not tied up with git, you can experiment without committing changes; if you don’t like them, just discard and build again. If your pipeline fails, fix the issue, build again and execution will resume from the crashing point.
Ploomber is robust to code style changes. It won’t trigger execution if you only added whitespace or formatted your source code.
Try it!
Click here to try out the live demo (no installation required).
If you prefer to run it locally:
pip install "ploomber[all]"
# create a new project with basic structure
ploomber new
ploomber entry pipeline.yaml
If you want to know how Ploomber works and what other neat features there are, keep on reading.
Inferring dependencies and injecting products
For Jupyter notebooks (and annotated Python scripts), Ploomber looks for a “parameters” cell and extracts dependencies from an “upstream” variable.
# annotated python file, it will be converted to a notebook during execution
import pandas as pd
# + tags=["parameters"]
upstream = {'some_task': None}
product = None
# +
df = pd.read_csv(upstream['some_task'])
# do data processing...
df.to_csv(product['data'])
In SQL files, it will look for and “upstream” placeholder:
CREATE TABLE {{product}}
SELECT * FROM {{upstream['some_task']}}
WHERE x > 10
Once it figured out dependencies, the next step is to inject products declared in the YAML file to the source code and upstream dependencies to downstream consumers.
In Jupyter notebooks and Python scripts, Ploomber injects a cell with a variable called “product” and an “upstream” dictionary with the location of its upstream dependencies.
import pandas as pd
# + tags=["parameters"]
upstream = {'some_task': None}
product = None
# + tags=["injected-parameters"]
# this task uses the output from "some_task" as input
upstream = {'some_task': 'output/from/some_task.csv'}
product = {'data': 'output/current/task.csv'}
# +
df = pd.read_csv(upstream['some_task'])
# do data processing...
df.to_csv(product['data'])
For SQL files, Ploomber replaces the placeholders with the appropriate table/view names. For example, the SQL script shown above will be resolved as follows:
CREATE TABLE clean
SELECT * FROM some_table
WHERE x > 10
Embracing Jupyter notebooks as an output format
If you look at the plot.py task above, you’ll notice that it has two products. This is because “source” is interpreted as the set of instructions to execute, while the product is the executed notebook with cell outputs. This executed notebook serves as a rich log that can include tables and charts, which is incredibly useful for debugging data processing code.
Since existing cell outputs from the source file are ignored, there is no strong reason to use .ipynb files as sources. We highly recommend you to work with annotated Python scripts (.py) instead. They will be converted to notebooks at runtime via jupytext and then executed using papermill.
Another nice feature from jupytext is that you can develop Python scripts interactively. Once you start jupyter notebook, your .py files will render as regular .ipynb files. You can modify and execute cells at will, but building your pipeline will enforce a top-to-bottom execution. This helps prevent Jupyter notebooks most common source of errors: hidden state due to unordered cell execution.
Using annotated Python scripts makes code versioning simpler. Jupyter notebooks (.ipynb) are JSON files, this makes code reviews and merges harder; by using plain scripts as sources and notebooks as products you get the best of both worlds: simple code versioning and rich execution logs.
Seamlessly mix Python and (templated) SQL
If your data lives in a database, you could write a Python script that connects to it, sends the query and closes the connection. Ploomber allows you to skip boilerplate code so you focus on writing the SQL part. You could even write entire pipelines using SQL alone.
jinja templating is integrated, which can help you modularize your SQL code by using macros.
If your database is supported by SQLAlchemy or it has a client that implements the DBAPI interface, it will work with Ploomber. This covers pretty much all databases.
Interactive development and debugging
Ploomber does not compromise structure with interactivity. You can load your pipeline and interact with it using the following command:
ipython -i -m ploomber.entry pipeline.yaml -- --action status
This will start an interactive session with a dag object.
Visualize pipeline dependencies (requires graphviz):
dag.plot()
You can interactively develop Python scripts:
dag['task'].develop()
This will open the Python script as a Jupyter notebook with injected parameters but will remove them before the file is saved.
Line by line debugging is also supported:
dag['task'].debug()
Since SQL code goes through a rendering process to replace placeholders, it is useful to see how the rendered code looks like:
print(dag['sql_task'].source)
Closing remarks
There are many more features available through the Python API that are not yet implemented in the spec API. We are currently porting some of the most important features (integration testing, task parallelization).
We want to keep the spec API short and simple for data scientists looking to get the Ploomber experience without having to learn the Python framework. For many projects, the spec API is more than enough.
The Python API is recommended for projects that require advanced features such as dynamic pipelines (pipelines whose exact number of tasks is determined by its parameters).
Where to go from here
Found an error in this post? Click here to let us know.



