Jupyter notebooks, you love them, or you hate them. The use of Jupyter notebooks is one of the most divisive topics in the data community. Of course, there are valid reasons to avoid notebooks, but I wish the conversation centered on the real problems instead of discussing already solved issues over and over again. In this blog post, I discuss the most common myths around notebooks and comment on the critical unsolved problems.
“You cannot version control notebooks”
Typically, notebooks are .ipynb files. Under the hood, .ipynb files are JSON files with a pre-defined schema, and they keep the code and the output in the same file. Standalone notebooks are convenient: you can work on a notebook, save it, come back to it tomorrow, and the results will be there. The caveat is that they are challenging to manage with git. First, .ipynb files blow up git repository size, and secondly, git diff (and GitHub Pull Requests) won’t work. This is how the diff view for a notebook looks like on GitHub:
Fortunately, we can easily fix this. There is an official JupyerLab extension that integrates git right into the Jupyter interface, allowing you (among other things), to diff notebooks. Here’s how it looks:
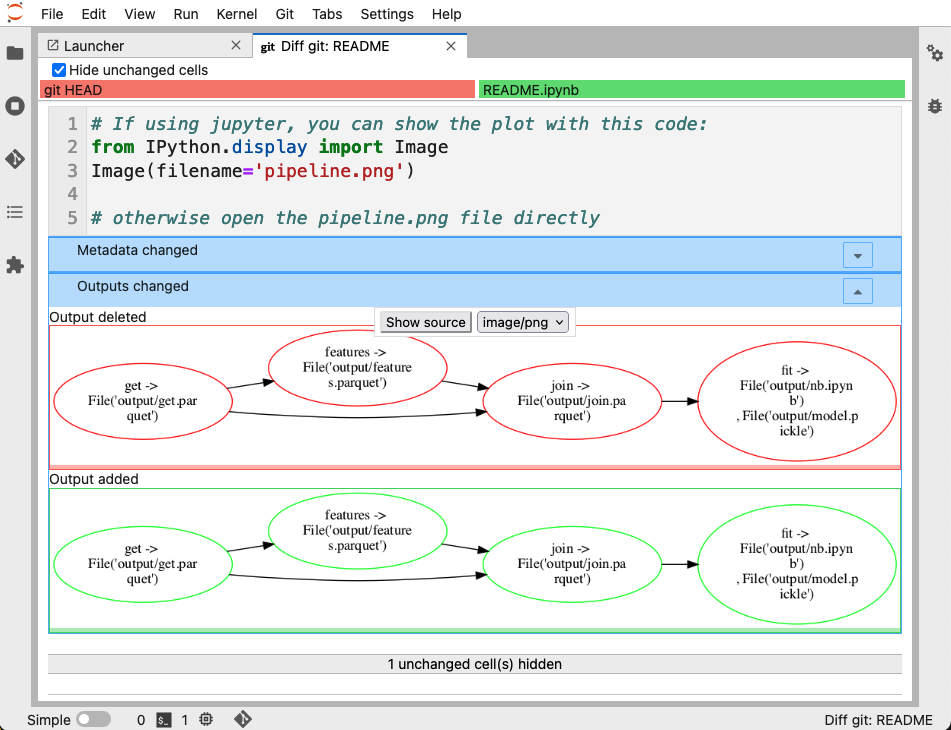
You can see that it clearly shows the difference, the current version on git has the graph with red borders, while the new one has green borders.
Another option is to use nbdime, which allows you to do the same from the terminal (in fact, the JupyterLab extension uses nbdime under the hood).
A third option (and my favorite!) is Jupytext. This package allows you to open .py files as notebooks. This way, you can edit your code interactively, but it’ll store a .py file once you save it.
When using Jupytext, you can use git diff and pull requests since your notebooks are simple scripts. The main caveat is that output is lost once you close the file, but you can use Jupytext’s pairing function, which stores the results in a separate .ipynb file, effectively storing source code and outputs two different files.
“You cannot test notebooks”
This one has a few angles. If we are thinking of testing notebooks as a whole, we can use papermill to execute them programmatically and evaluate their results. For example, you could write a test case with papermill like this:
import json
from pathlib import Path
import papermill as pm
def test_my_notebook()
# execute notebook
pm.execute_notebook('my-nb.ipynb', 'output.ipynb')
# ensure notebook creates model
assert Path('model.pickle').is_file()
# check it stores metrics
metrics = json.loads(Path('metrics.json').read_text())
assert list(metrics.keys()) == ['accuracy', 'precision', 'recall']
A simpler alternative is to run a smoke test (that is, checking that the notebook runs, but skip output checking). You may write a script like this to execute in your CI system (e.g. GitHub Actions):
papermill my-nb.ipynb output.ipynb
Finally, you may be interested in unit testing parts of the notebook (for example, a function defined inside the notebook). I recommend defining functions outside the notebook and importing them into the notebook. This way, you can unit test the functions as you would with any other function. However, if you want to write the function inside the .ipynb file, you can use testbook.
Testbook allows you to extract definitions from a .ipynb file and test them:
from testbook import testbook
@testbook('my-nb.ipynb', execute=True)
def test_func(tb):
# get function defined in the notebook
add_numbers = tb.get('add_numbers')
# test function
assert add_numbers(1, 2) == 3
“You cannot modularize notebooks”
Another criticism is that you cannot modularize notebooks. However, that’s straightforward with Ploomber. Say you have three notebooks (load.ipynb, clean.ipynb, and plot.ipynb), you can create a pipeline that executes them in order like this:
tasks:
- source: load.ipynb
product: outputs/load.ipynb
- source: clean.ipynb
product: outputs/load.ipynb
- source: plot.ipynb
product: outputs/load.ipynb
Then, you can run them with:
ploomber build
Ploomber will execute the notebooks in order and produce a copy each one, so you record outputs of this run.
Try it out!
# install ploomber
pip install ploomber
# download example
ploomber examples -n guides/first-pipeline -o example
cd example
# install dependencies
pip install -r requirements.txt
# run pipeline
ploomber build
Modularized notebooks open many possibilities: they are easier to collaborate, test, and are computationally efficient (you can run independent notebooks in parallel!). Therefore, we highly encourage you to write pipelines instead of big monolithic notebooks.
So, what are the real problems?
Hidden state

Given a notebook, we say it has a hidden state when executing it linearly returns different outputs than the stored ones. For example, the left notebook does not have hidden state, because if you restart the kernel and run all cells in order, you’ll get the same results (the number 3). But, the right notebook has hidden state: if we restart the kernel and run all cells in order, the final output won’t match (the recorded one is 42, but we’ll get 3). This problem is so pervasive that there’s even a Nature blog post about it. Hidden makes it impossible to reproduce our results.
We can partially solve this problem with testing: we can execute our notebooks linearly on each git push to ensure reproducible results. However, this does not solve the problem entirely, as we may still end up with hidden state during notebook development and find out until we push to the repository and the CI breaks.
A popular approach are reactive notebooks, which automatically re-compute cells to prevent hidden state. For example, say I have a notebook like this:

If I’m running a reactive notebook and edit the cell a = 1, the notebook will automatically run the third and fourth cells (total = a + b, and total) because they depend on the value of a, effectively updating and printing the new value of the total variable. Reactivity is a neat feature already available in Pluto, the notebook system for Julia, and it’s also available in some Jupyter commercial distributions. Fortunately, people are already working on an open-source solution for reactive kernels in Jupyter.
However, reactive kernels come with limitations. In many cases, some cells may be too expensive to re-compute, which may lead to user’s frustration if running a seemingly small computation suddenly triggers an hour-long calculation. To what extent reactive kernels should be enforced, or what alternatives we shoould build is a topic worth discussing.
Code quality
Code quality in notebooks is the problem that I find the most fascinating, as it always leads to perspectives that tackle the problem from different fronts. Code quality is by far the biggest problem with notebooks today.
Many argue that notebooks lead to poor code quality. Is that true? I don’t think so. Low-quality code exists everywhere; it’s not a Jupyter-exclusive problem. My take is that there is so much hard-to-read code in notebooks because Jupyter democratized computing. You no longer need to master an IDE and a terminal to develop data analysis code; you can open a notebook and get started right away. So all of a sudden, we have plenty of people with non-software backgrounds working as data scientists in all industries, and that is a fantastic thing. Everyone should have the opportunity to code and create value, and Jupyter is the entry door for many people to the world of computing.
The problem isn’t that notebooks produce low-quality code. As a community, we have failed to mentor those people who know a lot of statistics and Machine Learning but don’t have much experience coding. The pervasiveness of low-quality notebooks are a consequence of the lack of processes to develop and deploy data analysis code. Hardly any data team uses code reviews as part of their development process, so how do we expect us to be good mentors and help level up the skills of our colleagues?
Concluding thoughts
Notebooks are the best way to explore and analyze data, but they have problems. So I wish we could move on from the notebooks cannot be versioned conversation and start talking more about improving the notebook-based development and deployment process. At Ploomber, we firmly believe that maximizing the speed at which data teams deliver value has notebooks at its core. So we’re working hard to fix all the notebook problems to hit the sweet spot between the power interactivity and the reliability of a well-defined development and deployment process. Join our community to keep the conversation going.



