As we have shared with you before, there are several tools to run Jupyter notebooks on our computer. Some of these tools include certain functionalities that, beyond just executing our notebooks from the start to the end, these functionalities allow us to take advantage of the computer’s resources to even carry out parallel executions. In this post we will show you how you can run some notebooks in parallel and we will compare the execution times of some of these tools.
Why run notebooks in parallel?
First of all, we need to understand what advantages it has and in what cases it helps us to be able to run notebooks in parallel. Among some of the advantages that exist, we highlight the following:
- We can run different tasks or processes simultaneously by using multiple computing resources during parallel notebook executions.
- Parallel notebook executions will allow us to save time, allowing general process executions to be carried out in less time by executing independent processes simultaneously (as mentioned in the previous point).
- Parallel notebook executions can generally help us to keep a more organized code structure because of the requirements to be able to perform those executions simultaneously.
Now that we know some of the benefits, a natural interrogative would be to look for a practical use case where running notebooks in parallel would be useful, so let’s take a look at a use case.
Use case: Parallelizing ML model training
Let’s imagine that we are working on a data process in which the obtaining, cleaning and preparation of data has been previously carried out. We are now at a crucial point in our process: we need to fit a model (or more than one) to our data. Ideally, we have an idea of which models might work, however, what we would like to do is try different models (with different parameters) so that, based on the results obtained, we can choose the one that best fits our data.
A first approach might be to work in a Jupyter notebook, running the entire code from the start to the end of the notebook, adding the corresponding code to load and prepare the data, and later to model and evaluate the results for that data. Then, a copy of the notebook would be created to test with another model, and so on until you have n notebooks for the n different models to be tested. The entire process would look like this:
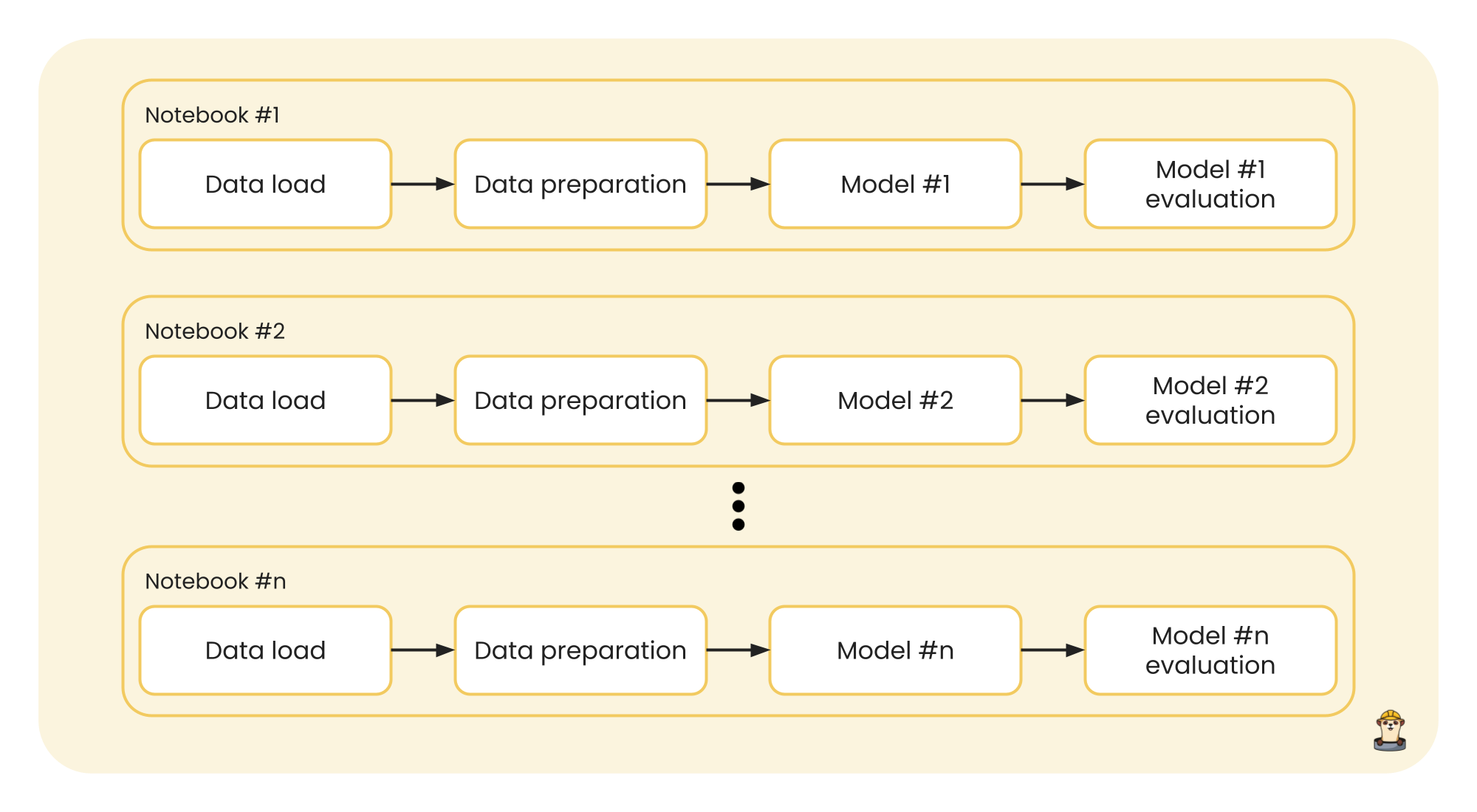
This process could be executed in parallel, since the notebooks are independent of each other and each one returns its own results. Furthermore, to add good code execution practices and a better work structure, since the data that enters a model is the same for each one, we can separate the data from the modeling and make our execution more efficient:
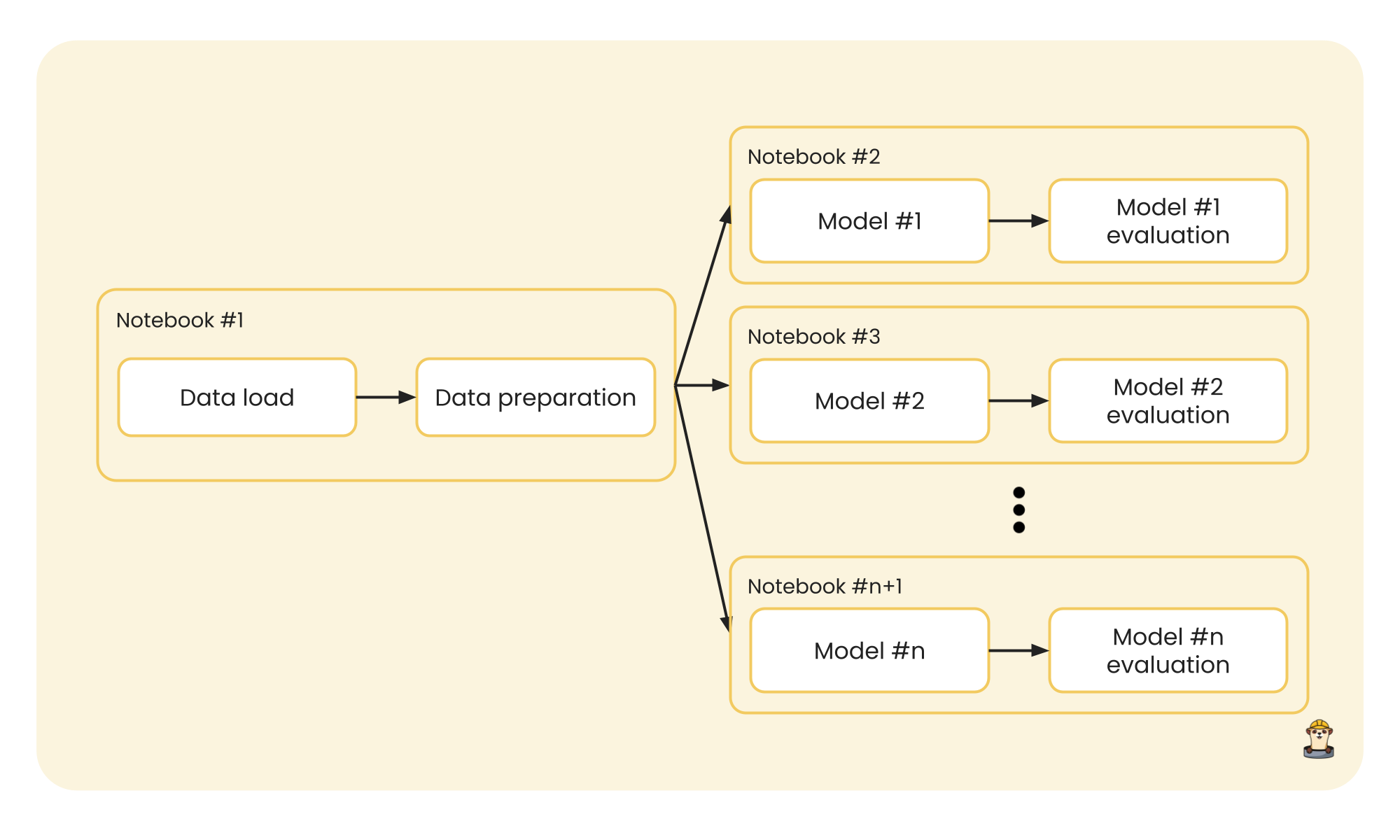
Again, running model training from a notebook for each model can be done in parallel to solve our problem in this use case.
As we have seen, running notebooks in parallel can be very useful, and even more so by adding a good work structure. Now it is time to know about some tools that will allow us to carry out these actions at a technical level.
Comparison of different tools
Some of the Jupyter notebook tools stand out for their versatility and simplicity of use to run an entire notebook, cell by cell. The extra benefits about some of these tools come when considering executions in parallel.
Here we will share the results after testing and evaluating some of these tools. Note that to make this comparison fair, it takes into account the use of the same code for all executions and we also use Python’s time module to measure the execution time. The notebooks used for benchmarking can be found here and correspond to the african_microbiome_portal_data repository. Serial execution cases (each notebook sequentially) are evaluated first, followed by parallel notebook execution cases.
Papermill
As a first option, we will use Papermill, which has a Python API that allows us to run different notebooks using some functions:
# This requires:
# pip install papermill
import papermill as pm
from glob import glob
for nb in glob('*.ipynb'):
pm.execute_notebook(
input_path=nb,
output_path=nb,
)
Execution time: 1:58.79 total
Ploomber serial
As a second option, we will use Ploomber with serial execution, which also has a Python API that allows us to execute different notebooks using the NotebookRunner function:
# This requires:
# pip install ploomber
from ploomber import DAG
from ploomber.products import File
from ploomber.tasks import NotebookRunner
from ploomber.executors import Parallel
from pathlib import Path
from glob import iglob
dag = DAG()
for path in iglob('*.ipynb'):
NotebookRunner(Path(path), File(path), dag=dag, papermill_params=dict(engine_name='embedded'))
if __name__ == '__main__':
dag.build(force=True)
Execution time: 59.324 total
Papermill with ploomber-engine
As a third option we will use Papermill again, but now with the ploomber-engine, which adds debugging and profiling features to Papermill:
# This requires:
# pip install papermill ploomber-engine
from glob import glob
for nb in glob('*.ipynb'):
pm.execute_notebook(
input_path=nb,
output_path=nb,
engine_name='embedded',
)
Execution time: 51.256 total
Ploomber in parallel (the fastest option!)
Finally, as a fourth option we use Ploomber again but now with its parallel notebook execution feature:
# This requires:
# pip install ploomber
from ploomber import DAG
from ploomber.products import File
from ploomber.tasks import NotebookRunner
from ploomber.executors import Parallel
from pathlib import Path
from glob import iglob
dag = DAG(executor=Parallel())
for path in iglob('*.ipynb'):
NotebookRunner(Path(path), File(path), dag=dag, papermill_params=dict(engine_name='embedded'))
if __name__ == '__main__':
dag.build(force=True)
Execution time: 25.440 total (the fastest option!)
Conclusion
As we could see, executing notebooks in parallel can be very useful because this allows us to carry out several independent processes at the same time and thus reduce processing time. To do so, there are different tools such as Papermill and Ploomber, which are similar in that each one has its Python API in order to be integrated into our workflows, but different in terms of performance as we have seen; being that the parallel executor of Ploomber allows the parallel execution of notebooks in record time. It should be noted that if you are more familiar with working with Papermill, ploomber-engine in its integration with Papermill gives it parallel execution capacity in a very good time too.
Even more advantages using Ploomber: Ploomber Cloud
Ploomber and plumber-engine can be run remotely in our servers via Ploomber Cloud, so you don’t need to worry about a robust infrastructure that can support your runs, we take care of it. You just need to prepare your pipelines with Ploomber and we do the magic for you!
Do you have a particular use case? We’d love to hear what you’re working on and help you. Send us a message through our Slack channel or drop an email to rodo@ploomber.io.




