In Ploomber 0.14, we introduced a new feature to allow users of editors such as VSCode and PyCharm to develop modular and maintainable pipelines using .py scripts. This blog post summarizes how this integration works.
Interactive .py files with the percent format
Jupyter is the most popular tool for developing Data Science projects; it offers an interactive environment to write literate programs that manipulate data. Furthermore, other editors and IDEs have embraced this idea with a few changes.
For example, Spyder introduced the percent format, which allows users to represent “notebooks” in .py files by splitting cells using a special # %% syntax, to represent each cell:
# %%
# first cell
x = 1
# %%
# second cell
y = 2
IDE tools such as VSCode and PyCharm have adopted the percent format since it provides a balanced experience between regular non-interactive .py scripts and .ipynb notebooks.
How our integration works
Ploomber helps users develop modular and maintainable pipelines, where each unit of work can be a script, notebook, or function. For example, a pipeline that uses two input datasets may look like this:
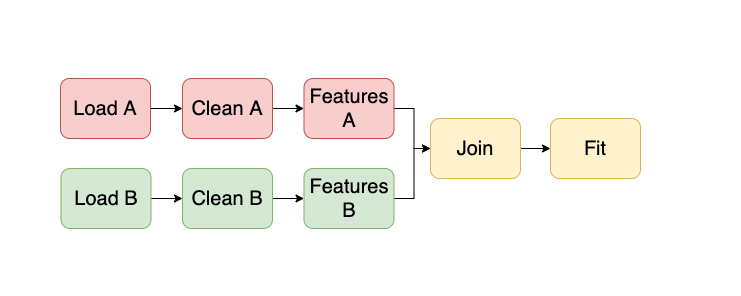
All tasks except those that load data, depend on upstream tasks; for example, Clean A uses Load A’s output as input. Therefore, our framework automatically injects a new cell when executing the pipeline from the command line. Hence, the Clean A task knows its input location (i.e., Load A’s output). So, for example, the Clean A script may look like this at runtime:
import pandas as pd
# %%
# declare load-a as an upsteram dependency
upstream = ['load-a']
# %%
# at runtime, Ploomber automatically injects this cell.
# this information is extracted from your pipeline declaration
upstream = {'load-a': 'path/to/data.parquet'}
# %%
# you can now load the data using the injected upstream variable
df = pd.read_parquet(upstream['load-a'])
However, one still wants to get that injected cell when developing interactively. Thanks to our Jupyter integration, the cell injection process happens automatically when the user opens a file, and we’re now bringing that functionality to other editors. With our new integration, users can inject the cell in all their scripts by running a single command:
ploomber nb --inject
After that, you can run your .py in percent format interactively, since it now includes the paths of the corresponding input - no hardcoding needed! Here’s a screenshot of VSCode after injecting the cell and running a .py in percent format:
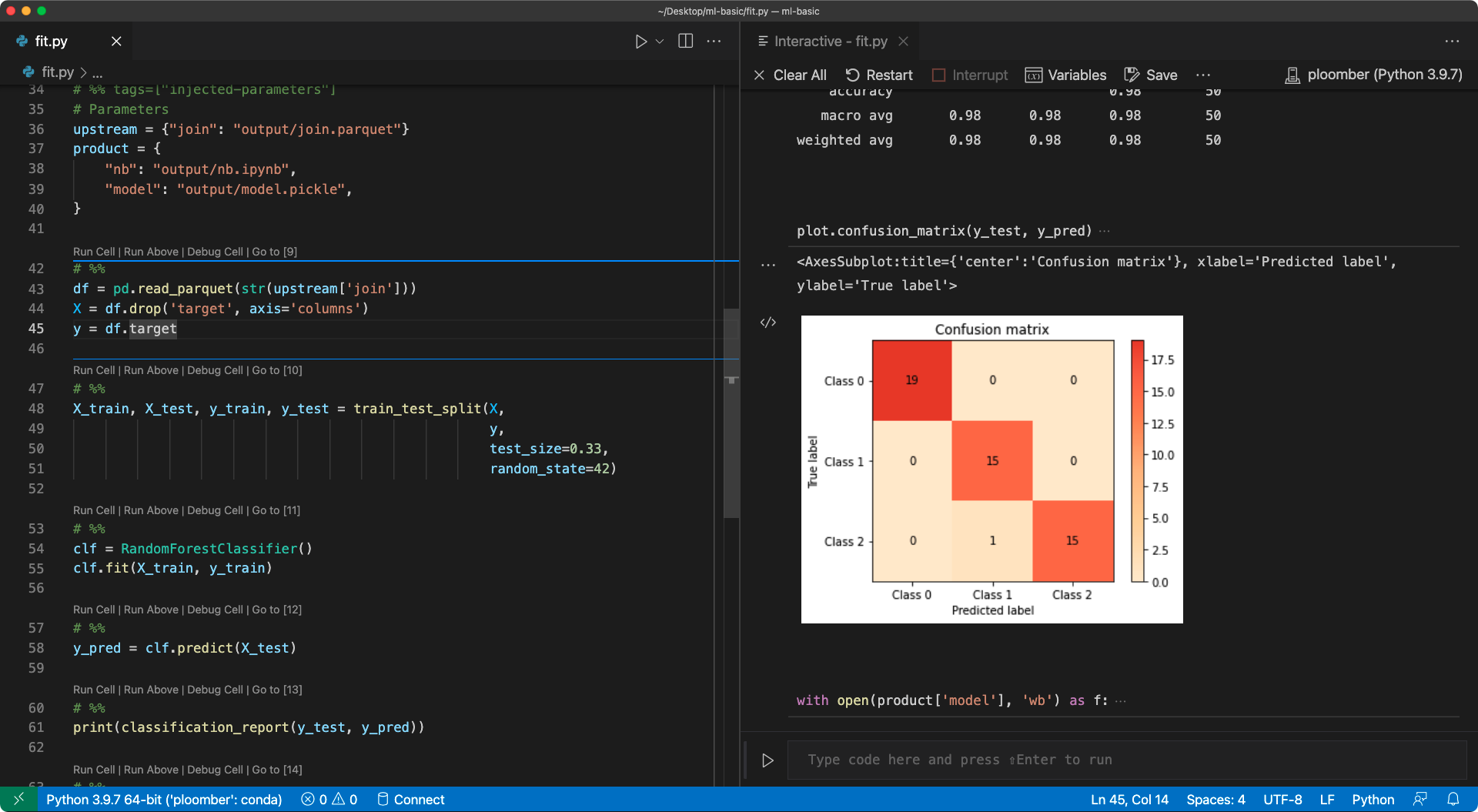
If you want to learn more, check out the user guide.
Get started
Get started in a few simple steps:
# install latest ploomber version
pip install ploomber --upgrade
# download example
ploomber examples -n templates/ml-basic -o example
cd example
# change scripts to the percent format
ploomber nb --format py:percent
# build pipeline
ploomber build
# inject cell
ploomber nb --inject
Now open fit.py and start running it interactively!
*You will need to make sure your IDE supports percent format
Closing remarks
The percent format is a great way to do data analysis interactively and keep the benefits of using .py files (like running git diff). In addition, by adopting Ploomber in your workflow, you can now break down your analysis logic in multiple steps to produce maintainable work and increase your team’s collaboration!
With this new integration, we want to support users from other editors and IDEs, so each team member can use whatever tool they like the most and collaborate smoothly; if you have any questions or suggestions, please join our community!



